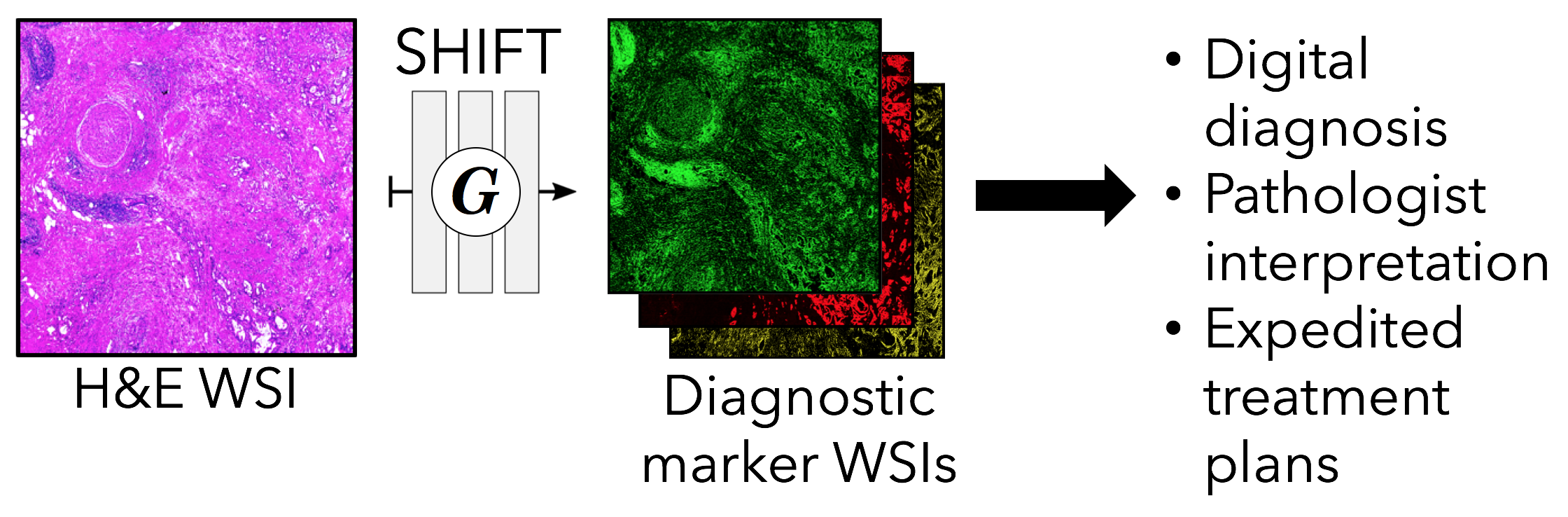
SHIFT: speedy histopathological-to-immunofluorescent translation of whole slide images using conditional generative adversarial networks
Abstract
Multiplexed imaging such as multicolor immunofluorescence staining, multiplexed immunohistochemistry or cyclic immunofluorescence enables deep assessment of cellular complexity in situ and, in conjunction with standard histology stains like hematoxylin and eosin (H&E), can help to unravel the complex molecular relationships and spatial interdependencies that undergird disease states. However, these multiplexed imaging methods are costly and can degrade both tissue quality and antigenicity with each successive cycle of staining. In addition, computationally intensive image processing such as image registration across multiple channels is required. We have developed a novel method, speedy histopathological-to-immunofluorescent translation (SHIFT) of whole slide images (WSIs) using conditional generative adversarial networks. This approach is rooted in the assumption that specific patterns captured in IF images by stains like DAPI, pan-cytokeratin, or alpha-smooth muscle actin are encoded in H&E images, such that a SHIFT model can learn useful feature representations or architectural patterns in the H&E stain that help generate relevant IF stain patterns. We demonstrate that the proposed method is capable of generating realistic tumor marker IF WSIs conditioned on corresponding H&E-stained WSIs with up to 94.5% accuracy in a matter of seconds. Thus, this method has the potential to not only improve our understanding of the mapping of histological and morphological profile into protein expression pro files, but also greatly increase the efficiency of diagnostic and prognostic decision-making.